Key master regulators of co-expressed genes
Using advantages of the design of the experiment allowing time-series style analysis, we identified master regulators and key-nodes (master regulators with positive feedback loop) from clusters of co-expressed genes and reconstructed diagrams of interactions between master regulators, transcription factors, and target genes for the top five master regulators [8].
We found that the main regulatory clusters were similar in both slow and fast muscles, but several unique patterns were discovered for each muscle type. In slow muscle, clusters were governed by regulators of proteasomal degradation and cell cycle regulators.
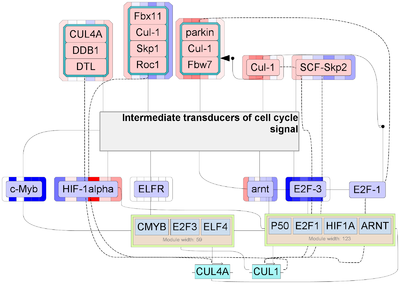
In particular, the most of regulators in slow muscle correspond to several E3 ubiquitin ligases (MAFbx/atrogin 1, Parkin) and components or regulators of SCF ubiquitin ligase complex (CUL1, SKP1, DDB1, ROC1, Ubs14) which are known signatures of disuse muscle atrophy in mammals as well as in humans [8,9]. In the fast muscle, proteasomal degradation and cell cycle regulators also played an important role, but the master regulators, transcription factors and target genes were different.
Unlike the slow muscle, calpain-2, for example, acts as one of the crucial master-regulators in fast muscles. Calpains are the members of calcium-dependent proteolytic system which along with ubiquitin-proteasomal system is an important player in muscle atrophy mechanism ((Taillandier et al. 1996; Murphy 2010; Huang and Forsberg 1998);
To summarize, using the atlas and time-series design of analysis we demonstrated that several genes that were known for their involvement in muscle biology act as master regulators in genetic response to atrophy.
Slow muscle (soleus)
Among key master-regulators that we identified from co-expressed genes clusters, genes encoded proteins involved in proteasomal degradation were overrepresented (Figure 6 ). In particular, the most of regulators in slow muscle correspond to several E3 ubiquitin ligases, among which there is a SCF family member Muscle atrophy F-box protein (MAFbx/atrogin 1). MAFbx together with another E3 ubiquitin ligase Muscle-specific RING finger protein 1 (MuRF1) are known signatures of disuse muscle atrophy in mammals as well as in humans [8,9]. In this cluster one of the most common key master-regulators is cullin-1 (CUL1). Cullin-1 is a major scaffold protein of SCF ubiquitin ligase complex, through SKP1 it binds to various F-box proteins, thus generating different types of muscle-specific ubiquitin ligases ( [36] [37]). In mammals CUL1 as well as other elements of SCF complex involve in ubiquitination of target substrates for subsequent degradation by 26S proteasome system ( [38,39];. Moreover, by ChIP-seq analysis it was shown that CUL1 indirectly interacts with DNA and can repress transcription of c-Myc-associated genes involved in RNA splicing and mitochondria regulation [40]. Among other master regulators SKP1 (S-phase kinase-associated protein 1) and DDB1 (DNA damage-binding protein 1) are adapter proteins providing binding cullin and F-box proteins thereby recognizing substrate for ubiquitination. ROC1 is also part of the SCF complex and appears as a regulator of cullin. In particular, ROC1 interacts with CUL1, indirectly promoting its nuclear accumulation by NEDD8 modification in HeLa cell culture [41]. Parkin is another ubiquitin-ligase which is involved in the elimination of defective mitochondria (mitophagy). Particularly, in skeletal muscles parkin plays an important role in maintenance of the mitochondrial function during endurance training [8,42].