Difference between revisions of "Enhancers"
(Created page with "We followed enhancers pipeline [15] to determine bidirectionally expressed permissive DPI1 clusters which were considered as enhancers. Less than 30% of fast muscle enhancers...") |
|||
| (2 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
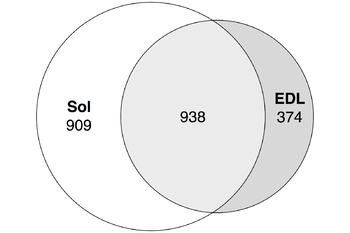
We followed enhancers pipeline [15] to determine bidirectionally expressed permissive DPI1 clusters which were considered as enhancers. Less than 30% of fast muscle enhancers were distinct (did not intersect) from slow muscles enhancers while roughly half of soleus enhancers were distinct from fast muscles enhancers (Figure 5). | We followed enhancers pipeline [15] to determine bidirectionally expressed permissive DPI1 clusters which were considered as enhancers. Less than 30% of fast muscle enhancers were distinct (did not intersect) from slow muscles enhancers while roughly half of soleus enhancers were distinct from fast muscles enhancers (Figure 5). | ||
| + | |||
| + | [[File:Enhancers - Figure5.png|350px|Enhancers - Figure5]] | ||
| + | |||
| + | <b>Figure 5.</b> Tissue-specific enhancers of soleus and EDL muscles Strandless coordinate intersection with 1nt cutoff. See [https://docs.google.com/spreadsheets/d/1C_zSjUghH_o1cgS93JspKfeK_aq1nW30fEaf0hlVP0M/edit?usp=sharing ST] for the full data. | ||
==Differentially expressed enhancers mediate muscle-specific transcription program== | ==Differentially expressed enhancers mediate muscle-specific transcription program== | ||
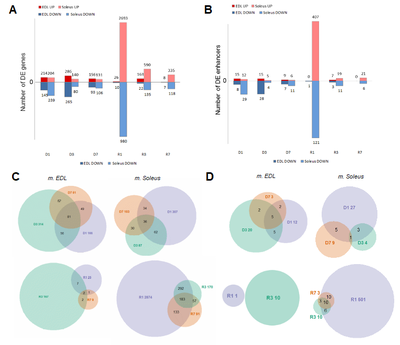
In both slow and fast muscles most of the enhancers were stably expressed in most of the phases of experiment, except for R1 where the slow muscles exclusively had approximately 30% of all its detected enhancers differentially expressed (Figure 3B). Such differential expression profile is consistent with the one for differentially expressed intergenic promoters and genes (Figure 3A). | In both slow and fast muscles most of the enhancers were stably expressed in most of the phases of experiment, except for R1 where the slow muscles exclusively had approximately 30% of all its detected enhancers differentially expressed (Figure 3B). Such differential expression profile is consistent with the one for differentially expressed intergenic promoters and genes (Figure 3A). | ||
| + | |||
| + | [[File:TSS and DEGs - Figure3.png|400px|TSS and DEGs - Figure3.png]] | ||
==Switch ON/OFF enhancers on first day of recovery (R1)== | ==Switch ON/OFF enhancers on first day of recovery (R1)== | ||
| − | We found that at least 26 enhancers in slow muscle were transcribed only on the first day of recovery (phase R1, see ST9). These 26 enhancers had 9 differentially expressed genes (AABR07004130.1, Stac2, Mn1, LOC102546495, Tsc22d2, AABR07044837.2, Pkdcc, Wnt1 and Ubash3b) in the reach of 1Mb, all of which appeared upregulated (see ST10). | + | We found that at least 26 enhancers in slow muscle were transcribed only on the first day of recovery (phase R1, see [https://docs.google.com/spreadsheets/d/1WUOkuY6yoW2PVqG4SZk5Wkvur8dzolDKxxHyGwuKUXk/edit?usp=sharing ST9]). These 26 enhancers had 9 differentially expressed genes (AABR07004130.1, Stac2, Mn1, LOC102546495, Tsc22d2, AABR07044837.2, Pkdcc, Wnt1 and Ubash3b) in the reach of 1Mb, all of which appeared upregulated (see [https://docs.google.com/spreadsheets/d/1X3ukg1mxqmTai99fjn2bBuegbuCSuZ1En-FxZ3z1NUY/edit?usp=sharing ST10]). |
In skeletal muscle, Stac proteins are essential for coupling membrane depolarization to Ca2+ release from the sarcoplasmic reticulum (SR). It was demonstrated that Stac proteins (1-3) interact with the II–III loop of CaV1.1, the principle subunit of Ca2+ channel, mediating the excitation–contraction in skeletal muscles [16]. | In skeletal muscle, Stac proteins are essential for coupling membrane depolarization to Ca2+ release from the sarcoplasmic reticulum (SR). It was demonstrated that Stac proteins (1-3) interact with the II–III loop of CaV1.1, the principle subunit of Ca2+ channel, mediating the excitation–contraction in skeletal muscles [16]. | ||
Latest revision as of 21:16, 2 March 2021
We followed enhancers pipeline [15] to determine bidirectionally expressed permissive DPI1 clusters which were considered as enhancers. Less than 30% of fast muscle enhancers were distinct (did not intersect) from slow muscles enhancers while roughly half of soleus enhancers were distinct from fast muscles enhancers (Figure 5).
Figure 5. Tissue-specific enhancers of soleus and EDL muscles Strandless coordinate intersection with 1nt cutoff. See ST for the full data.
Differentially expressed enhancers mediate muscle-specific transcription program
In both slow and fast muscles most of the enhancers were stably expressed in most of the phases of experiment, except for R1 where the slow muscles exclusively had approximately 30% of all its detected enhancers differentially expressed (Figure 3B). Such differential expression profile is consistent with the one for differentially expressed intergenic promoters and genes (Figure 3A).
Switch ON/OFF enhancers on first day of recovery (R1)
We found that at least 26 enhancers in slow muscle were transcribed only on the first day of recovery (phase R1, see ST9). These 26 enhancers had 9 differentially expressed genes (AABR07004130.1, Stac2, Mn1, LOC102546495, Tsc22d2, AABR07044837.2, Pkdcc, Wnt1 and Ubash3b) in the reach of 1Mb, all of which appeared upregulated (see ST10).
In skeletal muscle, Stac proteins are essential for coupling membrane depolarization to Ca2+ release from the sarcoplasmic reticulum (SR). It was demonstrated that Stac proteins (1-3) interact with the II–III loop of CaV1.1, the principle subunit of Ca2+ channel, mediating the excitation–contraction in skeletal muscles [16].
In the meantime, Wnt proteins are secreted ligands which bind to distinct receptors and activation of Wnt-mediated signaling pathway leads to GSK3 inhibition which, in turns, results in the muscle hypertrophy. Moreover, in vitro treatment of the myotubes with Wnt induces cell differentiation assuming an alternative way for the phenotype [17]. Pkdcc gene encoding protein kinase domain-containing protein, cytoplasmic has a more broad functional role in organogenesis via phosphorylation of extracellular proteins and endogenous proteins in the secretory pathway. It has only been shown in several genome wide studies that Pkdcc is significantly associated with the skeletal system development [17] [18].
Remarkably, data analysis has identified Mn1 which encodes a transcriptional activator and has a physiological role in cancer as an up-regulated gene during recover, while the gene transcript was in a top-list of genes downregulated in the skeletal muscle after both neuromuscular electrical stimulation and resistance exercise [19]. Moreover, recently published investigation of its physiological role has demonstrated Mn1 is higher expressed in fetal and adult skeletal muscle compared to other human organs and tissues and participates in the regulation of target genes through interaction with the transcription factors Pbx1, Pknox1, and Zbtb24 [20]. Taken together, the observed up-regulation of Mn1 expression in recovery stages can be explained by its pivotal role in developmental processes and cell differentiation [21] [22].
The identification of Ubash3b upregulation associated with corresponding enhancer switches also represents biologically meaningful outcome since the gene encodes a protein that was found to inhibit endocytosis of epidermal growth factor receptor (Egfr) [23]. Endocytosis is the general path for cellular receptors like Egfr to reduce the signaling response via negative feedback whereas Egfr-dependent signaling pathway promotes functional rescue of dystrophin-deficient satellite cells and enhances skeletal muscle regeneration and strength in mice [24].